Machine Learning Mastery Python Mini Course#
https://machinelearningmastery.com/python-machine-learning-mini-course/
import pickle
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
from pandas.plotting import scatter_matrix
from sklearn.preprocessing import StandardScaler
from sklearn.model_selection import KFold, cross_val_score, train_test_split
from sklearn.linear_model import LogisticRegression, Ridge
from sklearn.model_selection import GridSearchCV
from sklearn.neighbors import KNeighborsRegressor
from sklearn.ensemble import RandomForestClassifier
Creating DataFrames#
Create a simple DataFrame#
then load one in from a CSV at a URL
# Create a DataFrame from a NumPy array
myarray = np.array([[1, 2, 3], [4, 5, 6]])
rownames = ["a", "b"]
colnames = ["one", "two", "three"]
mydataframe = pd.DataFrame(myarray, index=rownames, columns=colnames)
mydataframe
| one | two | three | |
|---|---|---|---|
| a | 1 | 2 | 3 |
| b | 4 | 5 | 6 |
Create DataFrame from CSV URL#
this is the Pima Indians “Onset of Diabetes Dataset”
it’s about the characteristics of the Pima Indians
the final
"class"column is the class of diabetes0 meaning “no diabetes” and 1 meaning “has diabetes”
# Create a Pandas DataFrame from a CSV file from a URL
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.data.csv"
names = ["preg", "plas", "pres", "skin", "test", "mass", "pedi", "age", "class"]
data = pd.read_csv(url, names=names)
data
| preg | plas | pres | skin | test | mass | pedi | age | class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 763 | 10 | 101 | 76 | 48 | 180 | 32.9 | 0.171 | 63 | 0 |
| 764 | 2 | 122 | 70 | 27 | 0 | 36.8 | 0.340 | 27 | 0 |
| 765 | 5 | 121 | 72 | 23 | 112 | 26.2 | 0.245 | 30 | 0 |
| 766 | 1 | 126 | 60 | 0 | 0 | 30.1 | 0.349 | 47 | 1 |
| 767 | 1 | 93 | 70 | 31 | 0 | 30.4 | 0.315 | 23 | 0 |
768 rows × 9 columns
Examining the Data#
Describe#
# Statistical Summary
data.describe()
| preg | plas | pres | skin | test | mass | pedi | age | class | |
|---|---|---|---|---|---|---|---|---|---|
| count | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 | 768.000000 |
| mean | 3.845052 | 120.894531 | 69.105469 | 20.536458 | 79.799479 | 31.992578 | 0.471876 | 33.240885 | 0.348958 |
| std | 3.369578 | 31.972618 | 19.355807 | 15.952218 | 115.244002 | 7.884160 | 0.331329 | 11.760232 | 0.476951 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.078000 | 21.000000 | 0.000000 |
| 25% | 1.000000 | 99.000000 | 62.000000 | 0.000000 | 0.000000 | 27.300000 | 0.243750 | 24.000000 | 0.000000 |
| 50% | 3.000000 | 117.000000 | 72.000000 | 23.000000 | 30.500000 | 32.000000 | 0.372500 | 29.000000 | 0.000000 |
| 75% | 6.000000 | 140.250000 | 80.000000 | 32.000000 | 127.250000 | 36.600000 | 0.626250 | 41.000000 | 1.000000 |
| max | 17.000000 | 199.000000 | 122.000000 | 99.000000 | 846.000000 | 67.100000 | 2.420000 | 81.000000 | 1.000000 |
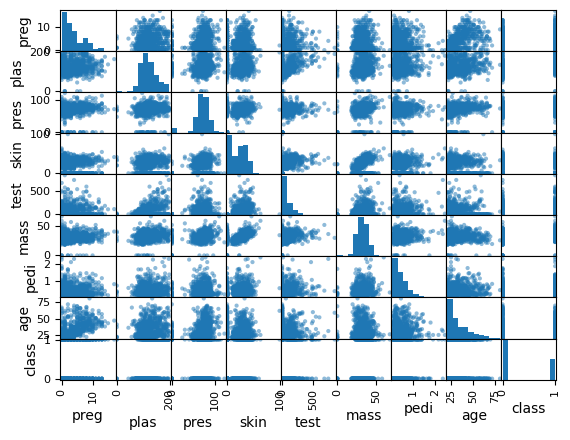
Scatter Plot Matrix#
shows each of the DataFrame columns combined with each other colum
“pairwise scatterplot”
where the data combines with itself, you get a histogram showing its distribution
in the other views, you get points plotted
so where
pregaligns withplasfor instanceif
pregis on the y-axis andplasis on the x-axisit plots the each point’s
(x,y)(plas,preg)
# Scatter Plot Matrix
scatter_matrix(data)
plt.show()
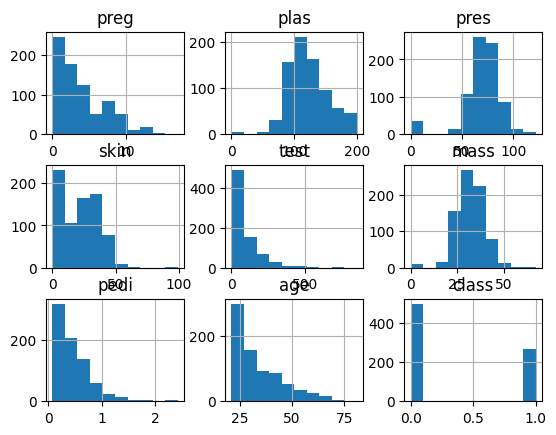
Histogram#
Shows the distribution of each of the columns
its those midline plots from the scatter plot matrix
data.hist()
array([[<Axes: title={'center': 'preg'}>,
<Axes: title={'center': 'plas'}>,
<Axes: title={'center': 'pres'}>],
[<Axes: title={'center': 'skin'}>,
<Axes: title={'center': 'test'}>,
<Axes: title={'center': 'mass'}>],
[<Axes: title={'center': 'pedi'}>,
<Axes: title={'center': 'age'}>,
<Axes: title={'center': 'class'}>]], dtype=object)
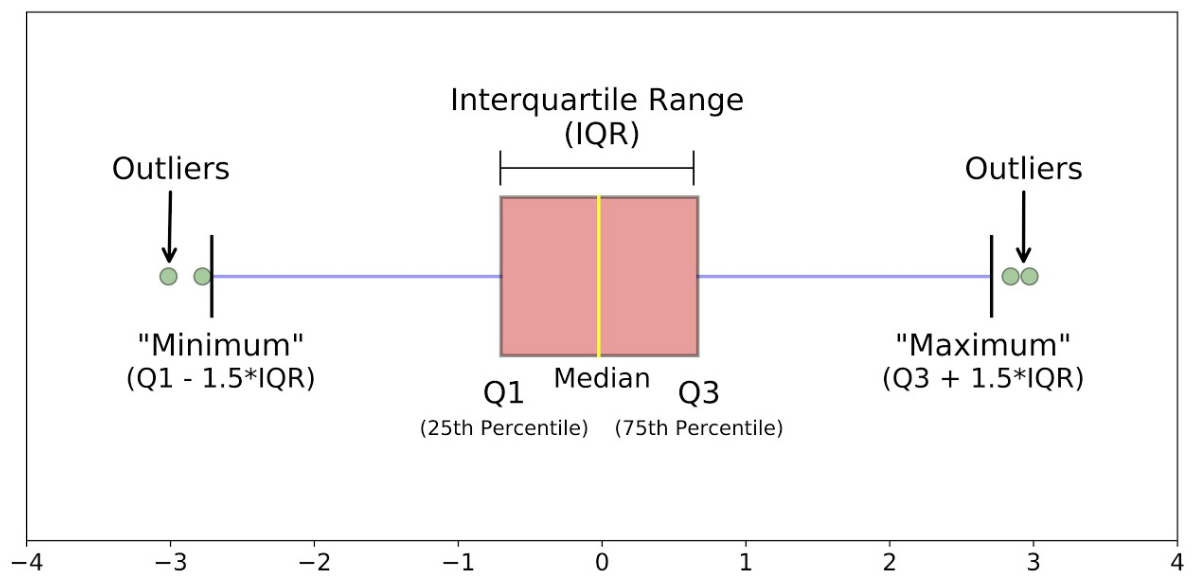
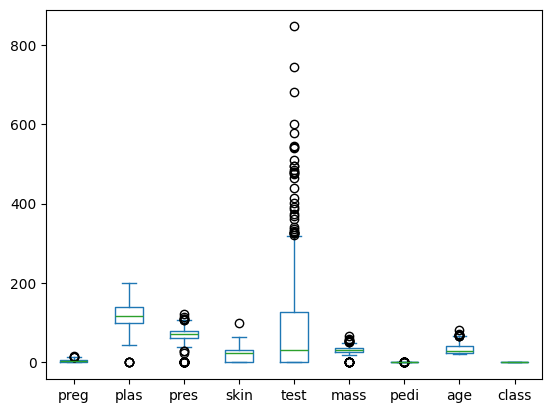
Box Plot#
data.plot(kind="box")
<Axes: >
Pre-Processing Data#
Standardize numerical data (set mean to 0, standard deviation of 1) with
scale and centeroptionsNormalize Numerical Data (min=1, max=1) using
rangeoptionAdvanced options like
Binarizing
Standardize (Scale and Center)#
mean = 0 , standard deviation = 1
so basically this?
hmm. Looks like image attachments don’t play well with html image links, though Stack Overflow seems to think it should work?
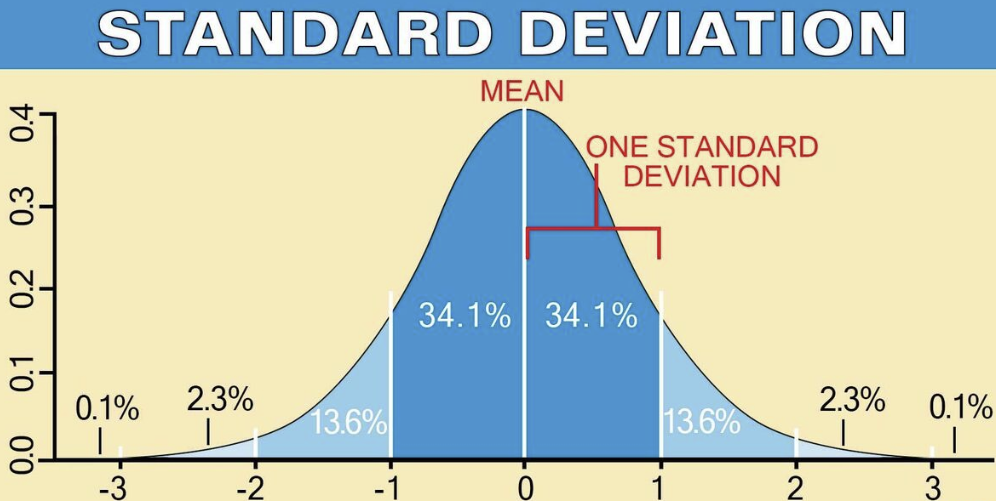
# Standardize data (0 mean, 1 stdev)
array = data.values
# separate array into input and output components
X = array[:, 0:8] # all rows, columns 0-7 (it's not inclusive of the end)
Y = array[:, 8] # all rows, column 8
scaler = StandardScaler().fit(X)
rescaledX = scaler.transform(X)
# set NumPy to print numbers with up to 3 decimal places
np.set_printoptions(precision=3)
# show rows 0-4 of the rescaled data
# sreveals that mean is now 1, the standard deviation of all of these is 1
rescaledX[0:5, :]
array([[ 0.64 , 0.848, 0.15 , 0.907, -0.693, 0.204, 0.468, 1.426],
[-0.845, -1.123, -0.161, 0.531, -0.693, -0.684, -0.365, -0.191],
[ 1.234, 1.944, -0.264, -1.288, -0.693, -1.103, 0.604, -0.106],
[-0.845, -0.998, -0.161, 0.155, 0.123, -0.494, -0.921, -1.042],
[-1.142, 0.504, -1.505, 0.907, 0.766, 1.41 , 5.485, -0.02 ]])
Resampling#
training dataset - data used to train the machine learning algorithm
you can’t use data that was used to train the algorithm to estimate the accuracy of the model for predicting on new data
Resampling - split the training dataset into subsets. Use some data to train, hold some back to check
K-Fold Cross Validation#
split the dataset into
10parts (9 to train, 1 to test)shuffle the order so the splits are randomized
set a random seed so that it’s reproducible
# create a K-fold cross-validator to split the dataset
kfold = KFold(n_splits=10, random_state=7, shuffle=True)
# create a logistic regression model
model = LogisticRegression(solver="liblinear")
# train the logistic regresssion model, use the kfold to split the data up
results = cross_val_score(model, X, Y, cv=kfold)
# print the accuracy and standard deviation of the model
print(f"Accuracy: {results.mean() * 100.0:.3f}")
print(f"Standard Deviation: {results.std() * 100.0:.3f}")
Accuracy: 77.086
Standard Deviation: 5.091
Evaluation Metrics#
many metrics can be used to evaluate how well an algorithm predicts on a dataset
Logloss#
results = cross_val_score(model, X, Y, cv=kfold, scoring="neg_log_loss")
print(f"Logloss: {results.mean():.3f}")
print(f"Logloss Standard Deviation: {results.std():.3f}")
Logloss: -0.494
Logloss Standard Deviation: 0.042
the following headings were mentioned in the chapter on the site
but there’s no further explanation or accompanying code
I guess I’m supposed to be figuring those out on my own?
there are probably equivalent ones on each other chapter that I missed
Confusion Matrix#
Classification Report#
RMSE and R-Squared#
Spot-Check Algorithms#
there are a LOT of machine learning algorithms
figuring out which one performs best on your data will require some level of trial-and-error
he refers to this as spot-checking algorithms
Linear Algorithms#
linear regression
logistic regression
linear discriminate analysis
Non-Linear Algorithms#
KNN - K-Nearest Neighbors
SVM - Support Vector Machine
CART - Classification and Regression Trees
start out with the Boston House Price dataset
# Prepare data from the Boston House Price dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/housing.data"
names = [
"CRIM",
"ZN",
"INDUS",
"CHAS",
"NOX",
"RM",
"AGE",
"DIS",
"RAD",
"TAX",
"PTRATIO",
"B",
"LSTAT",
"MEDV",
]
dataframe = pd.read_csv(url, delim_whitespace=True, names=names)
array = dataframe.values
X = array[:, 0:13]
Y = array[:, 13]
kfold = KFold(n_splits=10, random_state=7, shuffle=True)
/tmp/ipykernel_2243/190392337.py:19: FutureWarning: The 'delim_whitespace' keyword in pd.read_csv is deprecated and will be removed in a future version. Use ``sep='\s+'`` instead
dataframe = pd.read_csv(url, delim_whitespace=True, names=names)
# Train a K-Nearest Neighbors model on the Boston House Price dataset
model = KNeighborsRegressor()
scoring = "neg_mean_squared_error"
results = cross_val_score(model, X, Y, cv=kfold, scoring=scoring)
print(f"Negative MSE: {results.mean():<.3f}\nStd Dev: {results.std():<.3f}")
Negative MSE: -38.852
Std Dev: 14.661
Ensemble Algorithms#
random forest
stochastic gradient boosting
Model Comparison and Selection#
after spot checking a few algorithms, must compare performance
Compare linear algorithms to each other on a dataset.
Compare nonlinear algorithms to each other on a dataset.
Compare different configurations of the same algorithm to each other.
Create plots of the results comparing algorithms.
this example trains a couple of algorithms on the PIMA diabetes dataset and compares their accuracy
# Compare Algorithms
from pandas import read_csv
from sklearn.model_selection import KFold
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import LogisticRegression
from sklearn.discriminant_analysis import LinearDiscriminantAnalysis
# load dataset
url = "https://raw.githubusercontent.com/jbrownlee/Datasets/master/pima-indians-diabetes.data.csv"
names = ["preg", "plas", "pres", "skin", "test", "mass", "pedi", "age", "class"]
dataframe = read_csv(url, names=names)
array = dataframe.values
X = array[:, 0:8]
Y = array[:, 8]
# prepare models
models = [
("Logistic Regression", LogisticRegression(solver="liblinear")),
("Linear Discriminant Analysis", LinearDiscriminantAnalysis()),
]
# evaluate each model in turn
scoring = "accuracy"
kfold = KFold(n_splits=10, random_state=7, shuffle=True)
print("Accuracy and Standard Deviation for each model:")
# train and evaluate each model in turn
for name, model in models:
cv_results = cross_val_score(model, X, Y, cv=kfold, scoring=scoring)
print(f"{name}: {cv_results.mean():.3f} ({cv_results.std():.3f})")
Accuracy and Standard Deviation for each model:
Logistic Regression: 0.771 (0.051)
Linear Discriminant Analysis: 0.767 (0.048)
Improving Model Accuracy#
Parameter Tuning#
once you’ve identified an algorithm or two that works well, tune them to improve their performance
for example, by tuning the parameters to your specific dataset
this sounds like hyperparameter tuning
scikit-learn has a couple of way to search for good parameters
grid search (that you specify)
random search
the below example uses a grid search to set the parameters of a linear Ridge model trained on the Pima Indians Diabetes Dataset
grid search is expensive since it trains and checks every combination
# Grid Search for Algorithm Tuning
# create an array of alpha parameters to check
alphas_list = [1, 0.1, 0.01, 0.001, 0.0001, 0]
alphas = np.array(alphas_list)
# create a parameter grid dictionary
# where the key is 'alpha' and the value is the array
# this format allows you to search a grid of multiple parameters
param_grid = dict(alpha=alphas)
# create a ridge regression model
model = Ridge()
# create a grid search object to check all values of alpha
# this time, use 3-fold cross validation
grid = GridSearchCV(estimator=model, param_grid=param_grid, cv=3)
# train the model for each
grid.fit(X, Y)
# announce the best
# best score of 0.280? that's pretty low
print(f"alphas checked: {alphas_list}")
print(f"best score: {grid.best_score_:.3f}")
print(f"best alpha: {grid.best_estimator_.alpha:.3f}")
alphas checked: [1, 0.1, 0.01, 0.001, 0.0001, 0]
best score: 0.280
best alpha: 1.000
Ensemble Predictions#
Practice bagging ensembles with
random forest and extra trees algorithms
gradient boosting machine and AdaBoost algorithms.
Practice voting ensembles by
combining the predictions from multiple models together
the following code demonstrates using the Random Forest algorithm on the Pima Indians Diabetes Onset dataset
Random Forest is a “bagged ensemble of decision trees”
# make a KFold cross-validator to split it up
kfold = KFold(n_splits=10, random_state=7, shuffle=True)
# set parameters and create the Random Forest model
num_trees = 100
max_features = 3
model = RandomForestClassifier(n_estimators=num_trees, max_features=max_features)
# train and score the model (takes longer than the lineaer ones)
results = cross_val_score(model, X, Y, cv=kfold)
print(f"Random Forest Accuracy: {results.mean():.3f}")
Random Forest Accuracy: 0.773
Finalize and Save#
once you’ve got a tuned, well-performing model, finalize and save it
practice making predictions on new (unseen) data
save the model to file
I seem to recall there was a format other than pickle that was better for machine learning models
load it from file and score it
# split the dataset into train and test sets
# this time do it manually since we're training and scoring separately
X_train, X_test, Y_train, Y_test = train_test_split(
X, # unsplit inputs
Y, # unsplit outputs
test_size=0.33, # hold back 1/3 of the data for testing
random_state=7, # set a seed for reproducibility
)
# Fit the Logistic Regression model on the training data (2/3rds of the data)
model = LogisticRegression(solver="liblinear")
model.fit(X_train, Y_train)
# save the model to disk as a compressed "pickle" file
filename = "finalized_model.sav"
pickle.dump(model, open(filename, "wb"))
# some time later...
# load the model from disk
loaded_model = pickle.load(open(filename, "rb"))
# check how well the loaded model performs on the test set
result = loaded_model.score(X_test, Y_test)
print(f"Logistic Regression Accuracy: {result:.3f}")
Logistic Regression Accuracy: 0.756